CASE REPORT
Discovery of Influenza A (H7N2) in a Cat After Admission to an Animal Shelter: A Case Report
Elizabeth Roberts1, Carolyn Allen1*, Robin Brennen2, Aleisha Swartz1, Brenda Dines1, Francine Cigel3, Mary Lea Killian4, Beate Crossley5, David L. Suarez6, Mia Torchetti4, Christine Watson3, Sally Slavinski7, Kathy Toohey-Kurth3‡ and Sandra Newbury1
1Shelter Medicine Program, Department of Medical Sciences, School of Veterinary Medicine, University of Wisconsin-Madison, Madison, WI, USA; 2Animal Care Centers of NYC, New York, NY, USA; 3Wisconsin Veterinary Diagnostic Laboratory, University of Wisconsin-Madison, Madison, WI, USA; 4Department of Agriculture, National Veterinary Services Laboratories, Animal and Plant Health Inspection Service, U.S. Ames, Iowa, USA; 5California Animal Health and Food Safety Laboratory, University of California, Davis, Davis, CA, USA; 6Southeast Poultry Research Laboratory, U.S. National Poultry Research Center, Agricultural Research Service, U.S. Department of Agriculture, Athens, GA, USA; 7New York City Department of Health and Mental Hygiene, New York, NY, USA
Abstract
This case report describes the discovery of a low pathogenic avian lineage influenza A (H7N2) (A/feline/New York/16-040082-1/2016) infecting a cat in a shelter environment. Low pathogenic avian influenza virus H7N2 had previously circulated in poultry and farmed waterfowl and had reportedly been eradicated from live bird markets in 2006. Its appearance in a cat caused concern for the local cat population and had the potential to negatively impact the agricultural industry and human health. The first cat diagnosed presented to the shelter with no apparent clinical signs. Later, conjunctivitis developed and then upper respiratory congestion, which progressed to severe pneumonia that was unresponsive to treatment and characterized by dyspnea, tachypnea, and collapse. A second cat, who had entered the shelter 17 days earlier and had died at an emergency clinic, was later considered to be the index case. The second cat had similarly arrived with no apparent clinical signs, developed respiratory congestion leading to pneumonia with tachypnea and dyspnea, and tested positive for influenza A. Clinical consultation and diagnostic testing through multiple organizations identified the virus as an avian lineage H7N2 influenza virus.
Keywords: cat; influenza A; H7N2; animal shelter; diagnostic testing; respiratory disease; disease investigation; feline; avian influenza; feline influenza; influenza
Citation: Journal of Shelter Medicine and Community Animal Health 2023, 2: 61 - http://dx.doi.org/10.56771/jsmcah.v2.61
Copyright: © 2023 Elizabeth Roberts et al. This is an Open Access article distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), allowing third parties to copy and redistribute the material in any medium or format and to remix, transform, and build upon the material for any purpose, even commercially, provided the original work is properly cited and states its license.
Received: 21 July 2023; Revised: 11 August 2023; Accepted: 13 August 2023; Published: 3 October 2023
‡California Animal Health and Food Safety Laboratory, University of California, Davis, San Bernardino, CA, USA
Competing interests and funding: The authors declare no potential conflicts of interest.
Disclaimer. The findings and conclusions in this publication are those of the authors and should not be construed to represent any official USDA or U.S. Government determination or policy.
Correspondence: *Carolyn Allen Email: carolynallen@wisc.edu
Reviewers Julie Levy, Linda Jacobson
In December 2016, a cat at a New York City animal shelter tested positive for a low pathogenic avian lineage influenza A H7N2 (A/feline/New York/16-040082-1/2016).1 After arriving in apparently good health, the cat developed severe respiratory illness that led to a rapid decline in clinical condition followed by humane euthanasia. The infection and the virus were identified through a partnership of the shelter veterinary staff, University of Wisconsin Shelter Medicine Program (UWSMP), the Wisconsin Veterinary Diagnostic Laboratory (WVDL), the National Veterinary Services Laboratories (NVSL; Ames, IA), and the Southeast Poultry Research Laboratory (SEPRL). Whole genome sequencing was performed to further characterize the virus (A/feline/New York/16-040082-1/2016) as a live bird market (LBM) lineage H7N2, which was very closely related to a subset of LPAI H7N2 viruses primarily found in LBMs. The LBM lineage H7N2 avian influenza A viruses had been considered eradicated since 2006. Following the events highlighted in this case report, over 500 cats in New York shelters were quarantined and provided care in a temporary treatment facility. The full-scale collaborative response and apparent eradication of the virus have been partly described by previous publications.2–9
Influenza A viruses (Alphainfluenzavirus) can infect avian and mammalian species and have caused epizootics and pandemics for centuries.10 Influenza strains are classified by the surface proteins hemagglutinin (HA) (types 1–18) and neuraminidase (NA) (types 1–11).11 Further characterization is done by genome sequencing to determine the host species lineage. H7 is one of the avian influenza HA subtypes, along with H5, that can mutate into highly pathogenic avian influenza capable of causing severe disease in birds.12 The transition from low pathogenicity to high pathogenicity in poultry happens when specific mutations in the cleavage site of the HA protein occur, allowing for systemic spread in certain species of birds.13 Avian influenza viruses occasionally spill-over to exposed mammals, including humans, with a potential for a range of disease including a high case fatality rate.12 One variant of H7N9 emerged in China in 2013, initially as a low pathogenic virus that eventually mutated to being highly pathogenic in birds. Both the low and high pathogenic variants could infect people with 616 fatalities in 1,564 diagnosed cases (40% case fatality rate).12,14 Infections in poultry farm workers, and one immunocompromised patient without known poultry exposure, have been documented from the same LBM lineage H7N2 lineage that is reported here, so the potential for zoonotic and cross-species transmission associated with the finding of this subtype in a cat was of particular concern.15,16
The United States Department of Agriculture (USDA) has conducted surveillance for avian influenza viruses in the LBM system since the 1980s. Between 1994 and 2006, a specific lineage of LPAI H7N2 persisted in LBMs and caused outbreaks in commercial poultry.17–19 The USDA instituted a control program specific to the LBM system, and once measures for eradication were in place, this H7N2 lineage has not been detected in birds since April 2006.20 Thus, this discovery of a related virus in a cat was a significant finding from an agricultural, public health, and veterinary perspective.
In addition to the backdrop of poultry surveillance for avian influenza viruses, outbreaks of canine influenza virus (CIV) H3N2 first emerged in the United States in the Chicago area in March 2015.21 Initially, CIV H3N2 spread from the Chicago metropolitan area into neighboring Midwestern states. By November 2016, outbreaks of CIV H3N2 had been reported in many states across the nation.22,23 Because of this, vigilance was high for CIV H3N2 in shelters, particularly for dogs, and since CIV H3N2 had previously been reported to infect cats,24 UWSMP had been screening for feline infection in shelters as well.
The goals of presenting this case study are threefold: (1) to describe the process of discovering a novel influenza virus in a cat, (2) to describe the clinical signs and necropsy findings in the first cat known to be infected with an LBM lineage H7N2 avian influenza A virus (A/feline/New York/16-040082-1/2016), and (3) to connect the experience of this discovery to preventive practices in shelters.
Case Description
Animal Care Centers of NYC (NYCACC) is a registered nonprofit organization contracting with the city of New York to provide shelter admission to any animal presented at one of their shelter locations. During 2016, the shelter admitted 28,170 cats and dogs, including 19,241 cats, between their five locations in the boroughs of Brooklyn, Bronx, Manhattan, Staten Island, and Queens. The shelter locations in the Bronx and Queens boroughs were intake-only facilities, and the Brooklyn, Manhattan, and Staten Island facilities were utilized for both intake and housing. While the movement of cats between facilities was not uncommon, the two cats described were both admitted and housed only at the Manhattan shelter. At the time of this case, respiratory disease had been common within the cat population. Treatment and diagnostic testing for respiratory disease in cats was also common, including outside care at referral hospitals when warranted. All housing units were single-compartment and routinely filled with one or more cats. Additional cages were routinely utilized in hallways to house cats. The shelter veterinarian reported the population as over capacity, with 210 cats housed in the Manhattan shelter where these cases were discovered.
Cat A, a 12-year-old female spayed domestic short-hair cat, was admitted to NYCACC on 12th November 2016 (day 0). In her previous household, she had lived with an adult, one child, and two cats. Her owner reported her to be an indoor-only cat who had lived in that household for her lifetime. She was last seen by a veterinarian 2 years prior with no known health issues or injuries.
On physical examination at intake, day 0 (November 12th), cat A had been described as healthy and had received standard preventives, including Feline Viral Rhinotracheitis Calici Virus Panleukopenia (FVRCP) vaccination, indoxacarb for flea control (Activyl®, Merck Animal Health), and pyrantel pamoate as a dewormer (Figure 1). During the following 11 days, Cat A had in-kennel interactions with volunteers and staff, including in-kennel enrichment, a behavioral assessment, and rabies vaccination. She was moved to publicly accessible adoption housing on day 4 (16th November).
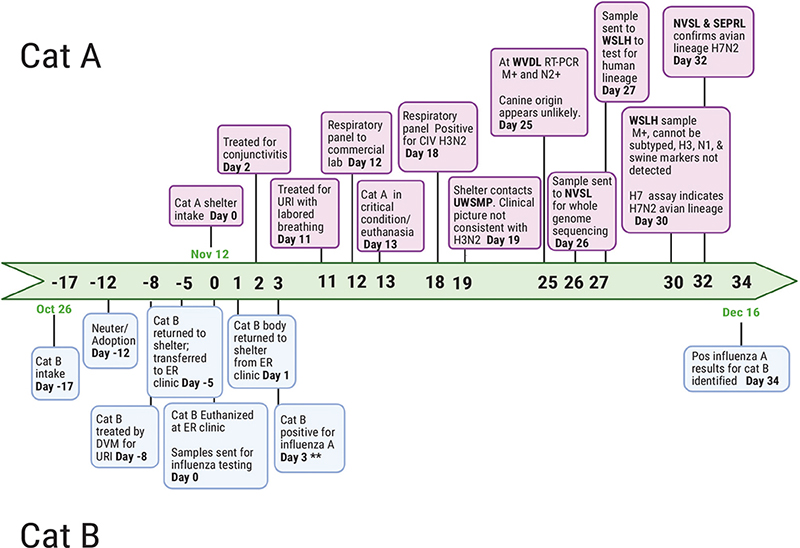
Figure 1. Timeline of events leading to the discovery of LBM origin H7N2 in a cat after admission to an animal shelter. The case material presented in this timeline occurred from October 26th to December 16th. **These results were reported but not found until the search for an index case (Cat B). ER: emergency clinic; UWSMP: University of Wisconsin Shelter Medicine Program; WVDL: Wisconsin Veterinary Diagnostic Laboratory; NVSL: National Veterinary Services Laboratories; Ames, IA; SEPRL: Southeast Poultry Research Laboratory; WSLH: Wisconsin State Laboratory of Hygiene. Created using Biorender.com.
Cat A had been examined again by the medical department on day 2 (14th November) for reports of ocular discharge and was noted by the veterinarian to have conjunctivitis with no respiratory component. She was started on Terramycin® (oxytetracycline hydrochloride, Zoetis) OU every 12 h. She later presented to the medical department on day 11 (23rd November) after developing respiratory signs, including congestion with no nasal discharge and normal bronchovesicular sounds. Doxycycline at 10mg/kg PO every 24 h was prescribed.
Two days later, on the morning of day 13 (25th November), Cat A was found in lateral recumbency open mouth breathing. On examination, she was tachycardic and dyspneic with abdominal effort and harsh lung sounds with wheezing. On a right lateral radiograph, she had an alveolar pattern with air bronchograms. Due to sudden clinical decompensation with suspected pneumonia, her prognosis was considered poor, and euthanasia was recommended and performed on day 13 (25th November).
Due to the unusual severity and rapid progression of Cat A’s pneumonia, swab samples were collected and sent for respiratory pathogen screening. Initial nasal swab samples sent to a commercial laboratory were tested accidentally on a canine respiratory panel. Positive results for H3N2 canine influenza were reported on day 18 (30th November). On day 19 (1st December), UWSMP was consulted because of the H3N2 results. This laboratory finding was unusual in cats and seemed improbable since H3N2 canine influenza was not clinically evident in dogs in the shelter. For these reasons, further diagnostic investigation was pursued.
A swab sample and Cat A’s body were both received by WVDL on day 25 (7th December). Using a multiplexed real-time PCR panel,21 Cat A was screened for influenza A, Chlamydia spp., feline coronavirus, feline calicivirus, feline panleukopenia virus, and Mycoplasma felis on day 25 (7th December). Of the screened pathogens, only influenza A was detected. Necropsy results further supported the diagnosis of influenza. On gross examination, hemorrhage mixed with fluid was noted from the nose and mouth (Figure 2a). The lungs were dark red, soft, and floated in formalin (Figure 2b and 2c). Microscopically, there was evidence of severe and acute bronchiolar denudation with airway remodeling and associated acute necrohemorrhagic interstitial pneumonia with pulmonary edema. Small, multifocal areas of lymphoid aggregates were present in pharyngeal lamina propria. Mycoplasma spp. was isolated on culture from nasal and lung swabs; no other significant growth occurred on bacterial culture, and fecal floatation yielded no parasite ova or oocysts.
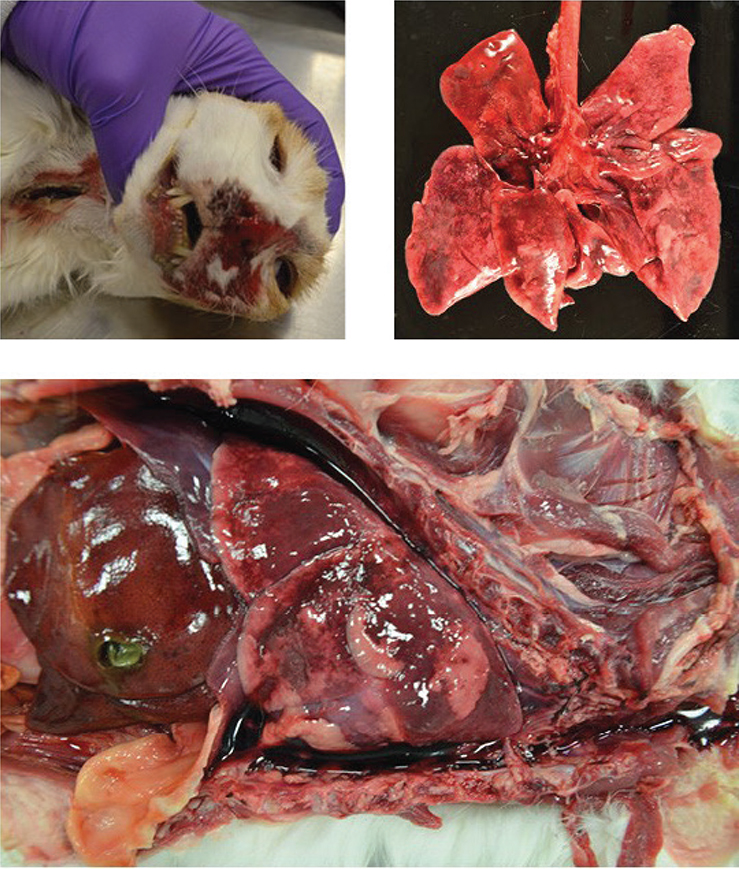
Figure 2a-c. Gross images from the necropsy of cat A. a) Hemorrhage mixed with fluid from nose and mouth. b) Multifocal to coalescing dark red lung lobes. c) Lung lobes (in situ) with multifocal to coalescing dark red areas and blood-tinged, watery fluid in the thoracic cavity.
Later, on day 34 (16th December), additional influenza A test results were identified in a kitten (Cat B) from the same shelter location. Cat B had a positive quantitative reverse transcription polymerase chain reaction (RT-qPCR) result for influenza A from the same commercial laboratory (sampled on day 0, 12th November). Influenza A results had been reported post-mortem for Cat B but were not noted until a search for previous cases was underway. No other reports of influenza A-positive feline cases were discovered before Cat B (Figure 1).
Cat B was initially found outside and brought to the shelter from the Bronx on Day 17 (26th October), 16 days before Cat A’s intake. He was described as healthy on intake examination and was vaccinated for FVRCP, dewormed with pyrantel pamoate, and received indoxacarb (Activyl®, Merck Animal Health) for flea control. Cat B received an in-kennel behavioral assessment on day 14 (29th October). He was neutered and adopted on day 12 (31st October).
On day 8 (3rd November), 8 days after his intake and 3 days after his adoption, Cat B was presented to a private veterinarian for an upper respiratory infection and was treated for 3 days; records were unavailable. Because his illness was progressing and not resolving, Cat B was re-admitted to NYACC on day 5 (7th November) to pursue more advanced treatment options. On examination, Cat B was congested with tachypnea and increased respiratory effort with crackles on auscultation. Due to difficulty with handling, a thoracic radiograph was not obtained. Cat B was diagnosed with suspected pneumonia and transferred to an emergency veterinary clinic, also on day 5 (7th November), 5 days before Cat A’s intake. At the emergency clinic, he received oxygen supplementation and supportive care. Despite treatment, he remained oxygen dependent and was subsequently euthanized due to declining clinical condition on day 0 (12th November). Post-mortem diagnostic testing at the emergency clinic included bacterial culture of lung tissue and RT-qPCR for influenza A from a commercial laboratory. As with cat A, cat B’s respiratory panel was inadvertently run as a canine sample. On day 3 (15th November), results from those samples reported bacterial culture was negative for aerobic and anaerobic growth, and the RT-qPCR was positive for influenza A, but no typing was reported.
These results had been reported by the laboratory post-mortem to the referral hospital who forwarded the results by fax to the shelter. Faxed results were initially mixed in a stack of papers and not received by shelter medical staff or added to the patient’s record. Results for cat B had not been reported to the New York City Department of Health or USDA at the time.
Later, when requested as part of a search for previous cases, the laboratory noted they were unable to further subtype the virus due to insufficient sample quantity. Cat B’s body had been returned to the shelter on day 1 (13th November) and was disposed of before he was identified as a suspect case on day 34 (16th December) (Figure 1).
Review of cat B’s history revealed he was found within one city block of an LBM. The LBM was part of the network of markets that undergo routine USDA surveillance for H5 and H7 influenza strains. During June and July 2016, LBMs in NY, NJ, and PA were subject to enhanced surveillance following detection of an unrelated LPAI H5N2 virus. The LPAI H7N2 LBM lineage had not been detected since 2006 even though routine surveillance has continued. Within the NYACC shelter system, all avian species present were isolated and tested, and all had negative results for influenza A.
Following the identification of H7N2 in cats A and B, an intensive investigation continued to identify new cases. A quarantine facility was established to house infected and exposed cats. While the virus caused significant morbidity, mortality was rare. The outbreak was determined to be over, and the H7N2 strain eradicated in March 2017. Some aspects have already been documented in lay and peer-reviewed publications.2–9
Diagnostic Testing
Following cat A’s euthanasia, a nasal swab and a lung sample were sent to WVDL and tested for influenza A, using a matrix-specific assay designed to be broadly reactive for diverse lineages of influenza.21 Matrix positive samples were further tested by RT-qPCR targeting the NA. The WVDL N RT-qPCR panel (N1 through N9) was designed to detect the N type regardless of species; however, to better detect canine H3N2, the N2 assay was modified to be more specific to the canine lineage. Because clinical information from the shelter population indicated that canine influenza H3N2 was unlikely to be the causative pathogen, the NA was further investigated. Both the general all-species N2 assay and the canine-specific N2 assay were performed. Discordant results revealed minimal reaction for the canine-specific N2 assay, while the all-species N2 assay reacted at a similar level as the matrix target reacted. This supported the clinical impression that the strain was likely not canine influenza H3N2. The NVSL (Ames, IA) was contacted to collaborate because of their ability to rapidly perform whole genome sequencing, and the sample was sent on day 26 (8th December).
On day 27 (9th December), a sample was also sent to Wisconsin State Laboratory of Hygiene (WSLH) to be tested with a Center for Disease Control (CDC) assay specific for human disease lineage. By day 30, the WSLH determined that the sample was positive for influenza A, but a subtype could not be identified. Testing for human seasonal H3, pandemic H1, and influenza A swine markers was all negative.
Also on day 30 (12th December), the gene-specific USDA National Animal Health Laboratory Network (NAHLN) H7 assay revealed that the H type was H7, completing the subtyping as H7N2. On day 32 (14th December), the whole genome sequence results analyzed by the NVSL and SEPRL identified LPAI H7N2 consistent with a lineage that was eradicated from the LBMs in 20061,20; specifically, the sequence was closest to LBM strains from the early 2000s. Once the pathogen was clearly identified as LPAI H7N2, which has the potential to become both a high consequence and zoonotic pathogen, the SEPRL, CDC, and multiple state and local public health agencies began a collaboration with UWSMP and WVDL to determine how to best respond.2–8
Although initially cat A had been reported as CIV H3N2 positive, only the N2 determination had been the result of testing, and the H3 was assumed because of the recent emergence and prevalence of H3N2 in the canine population in the U.S. Analysis of the virus revealed H7N2 supporting initial clinical impressions from the shelter population that H3N2 was unlikely to be present in the shelter.
Discussion
Focusing on the process of discovery of this novel virus in cats housed in a shelter setting illuminates the importance of having a plan for responding to infectious disease in animal shelters as well as laboratories and private practice. Monitoring frequency and severity of infectious disease, as well as having a system to respond if severe disease or a new disease trend presents, is critical. Investigation of etiology is indicated when clinical signs are severe, cases are not responding to treatment as expected, or a zoonotic condition is suspected.25 An avian influenza virus newly infecting cats in a shelter was a substantial concern because of the potential for widespread dissemination not only in the shelter population but also in the larger metropolitan area. Early recognition and response to unusual disease, along with rapid identification of the agent, helped prevent widespread infection outside the shelter environment. Figure 3 illustrates the key points of a systematic plan that would guide response to any unusual infectious disease event. This figure could be used proactively to develop plans and procedures for infectious disease response (Figure 3).
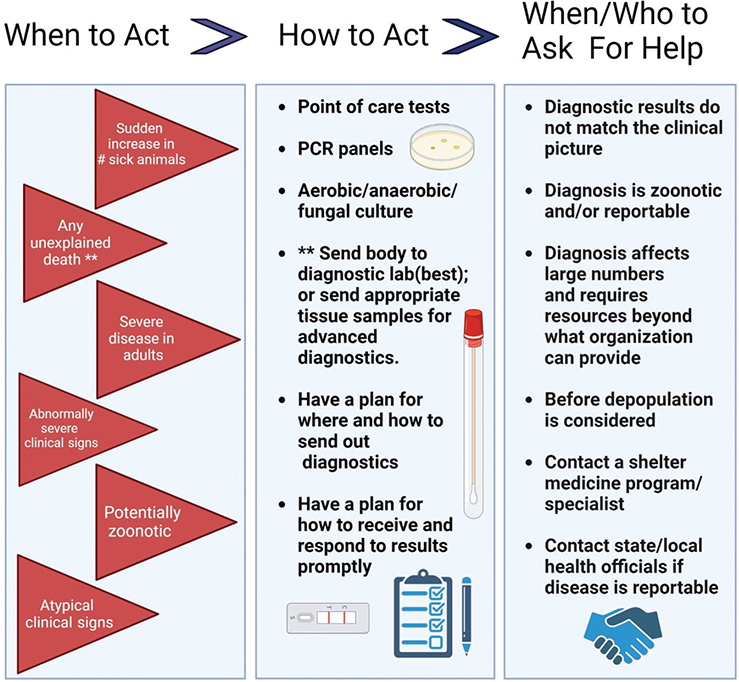
Figure 3. Clinical infectious disease management flow chart. Panel A depicts red flag situations that should raise suspicion for infectious disease. Even if only seen in one animal, these red flag situations may pose increased risk of morbidity and mortality in a shelter population. Panel B depicts diagnostic options that should be considered in response to the red flags in panel A. Panel C describes scenarios where outside consultation with shelter medicine experts is advisable. Created using Biorender.com.
The absence of unusual disease in dogs was equally critical information in identifying the etiology of disease in this outbreak. When CIV H3N2 infects dogs in a shelter, illness typically spreads in a matter of days. The population of dogs who are sick will balloon in a dramatic and often unmistakable way.22 This wave-like infection of dogs with respiratory disease was not occurring at the time of cat A’s illness. The diagnosis of H3N2 in a cat coupled with this lack of clinical disease in the dogs in the shelter prompted UWSMP to rapidly seek further testing and subtyping of samples from cat A to determine if a novel influenza virus might be present (Figure 3).
Finding previous cases provided context for the timeframe of the outbreak and where transmission may have occurred. An effort was made to identify previous cases by reviewing records of previously sick cats and historical results of respiratory testing. Shelter-acquired infection was a concern because of cat A’s history and her duration of stay in the shelter before clinical presentation of severe respiratory disease. While identifying cat B as the earliest recognized case did provide a longer, more complete retrospective contact tracing time frame, cat A and cat B did not overlap in the shelter, suggesting infection was likely not transmitted directly between these two cats.
Investigation of the virus would have begun even 15 days earlier if cat B’s results had been noted at the time results were first sent out. While procedures were in place for submitting diagnostic testing from animals with unusual or severe disease, practices for receiving and reviewing test results allowed for a lapse in recognition and delays in required reporting. Requirements from the New York City Department of Health in 2016 included reporting any unusual disease in an animal within 24 h and immediate reporting for novel influenza.26 Reporting requirements have not changed substantially since 2016.27 Multiple professional entities are listed as possible reporters (e.g., veterinarians, animal hospitals, animal shelters, and veterinary diagnostic laboratories), which may have left a lack of clarity for who was ultimately responsible.
The identification of influenza is based on the detection of the readily conserved influenza A matrix gene (M).28 Once influenza A is detected, then subtyping is performed to determine HA and NA subtypes. Sequence analysis provides the species lineage. When cat A’s sample was tested, an important point of confusion at the subtyping step occurred because the cat’s sample was initially processed as canine. In November 2016, the commercial laboratory’s canine respiratory panel included testing for H1N1, H3N2, and H3N8. When the N2 determination was made, the H3 was assumed since H3N2 was known to be in circulation. In this case, when more complete typing was done, the virus was determined to be H7N2 and closely related to the avian lineage H7N2 from the LBM, dramatically changing the response.1
Necropsy and histology results from cat A supported that clinical signs were related to infection with an influenza virus. Hemorrhage and edema with denuding of respiratory epithelium have been characteristic findings of canine influenza A infection in companion animals. Mycoplasma may have played a secondary role in increasing the severity of cat A’s pneumonia and clinical decline.29,30
Correct determination of etiology had profound implications because the continued circulation of an LPAI H7N2 that infects cats could have had a dramatic effect on feline health and welfare, with possible repercussions for public health and animal agriculture. If this LPAI H7N2 had become a new circulating virus of cats in the United States, the population of cats and in-contact humans or other animals might have provided opportunities for exposure, mutation, and re-assortment of the virus.31 Identification of the virus, which had been thought to be eradicated, raised concerns, especially that H7N2 might make its way back into poultry populations in the United States. Through mutation and re-assortment, influenza A viruses have demonstrated the ability to cross the species barrier and to establish continuous infections in new hosts.31 Cats have exposure to people and other animals both in shelters and in the community, providing opportunities for cross-species transmission events to occur.32,33
Conclusions
Identification of this novel influenza virus in cats was facilitated by clinical vigilance, initiated by clinical interpretation of diagnostics, and clarified by the use of a combination of a broadly reactive matrix assay followed by a broadly reactive NA typing assay. Confirmation was accomplished through full genome sequencing. Identification was delayed by errors in species identification (cat vs dog) in two separate submissions, initial incomplete identification of the virus (cat B), assumptions about the HA component of the virus (cat A), and poor communication and processing of incoming lab results.
This case highlights several points in response planning for influenza and other infectious agents. Very specific tests may miss novel agents or known viruses when mutations have occurred. Typing influenza-positive samples and characterizing virus is a common practice in the agricultural sector. Even within an established outbreak in companion animals, it is prudent to type and characterize the first influenza-positive sample for each new emergence or new species affected. In addition, diagnostic testing is best used in conjunction with clinical interpretation to arrive at a diagnosis. Systems for receiving and reviewing results are crucial, and follow-up to check for results should be integral to the submission process.
Multiple organizations and agencies collaborated closely to rapidly identify and confirm the etiology of disease. The possibility of widespread infection in cats created concerns ranging from impacts on feline health and welfare to potential zoonotic transmission and grave agricultural consequences.
Acknowledgments
Appreciation is noted for the generous funding provided by Maddie’s Fund in support of this outbreak response. Appreciation is also noted for the many hours spent by the USDA, CDC, New York State Agriculture, and New York State and New York City Public Health officials in response to this outbreak. In addition, a special acknowledgment is given to the dedicated response contributed by employees and volunteers at the New York City Animal Care Center, the Wisconsin State Veterinary Diagnostic Laboratory, and the University of Wisconsin–Madison Shelter Medicine Program.
References
| 1. | Newbury SP, Cigel F, Killian ML, et al. First Detection of Avian Lineage H7N2 in Felis catus. Genome Announc. 2017;5(23): 1–2. doi: 10.1128/genomeA.00457-17 |
| 2. | Lee CT, Slavinski S, Schiff C, et al. Outbreak of Influenza A(H7N2) among Cats in an Animal Shelter with Cat-to-Human Transmission—New York City, 2016. Clin Infect Dis. 2017;65:1927–1929. doi: 10.1093/cid/cix668 |
| 3. | Belser JA, Pulit-Penaloza JA, Sun X, et al. A Novel A(H7N2) Influenza Virus Isolated from a Veterinarian Caring for Cats in a New York City Animal Shelter Causes Mild Disease and Transmits Poorly in the Ferret Model. J Virol. 2017;91:e00672-17. doi: 10.1128/JVI.00672-17 |
| 4. | Marinova-Petkova A, Laplante J, Jang Y, et al. Avian Influenza A(H7N2) Virus in Human Exposed to Sick Cats, New York, USA, 2016. Emerg Infect Dis. 2017;23:2046–2049. doi: 10.3201/eid2312.170798 |
| 5. | Poirot E, Levine MZ, Russell K, et al. Detection of Avian Influenza A(H7N2) Virus Infection Among Animal Shelter Workers Using a Novel Serological Approach—New York City, 2016–2017. J Infect Dis. 2019;219:1688–1696. doi: 10.1093/infdis/jiy595 |
| 6. | Blachere FM, Lindsley WG, Weber AM, et al. Detection of an avian lineage influenza A(H7N2) virus in air and surface samples at a New York City feline quarantine facility. Influen Other Respir Viruses. 2018;12:613–622. doi: 10.1111/irv.12572 |
| 7. | Jain S, Murray EL. The Cat’s Meow: Using Novel Serological Approaches to Identify Cat-to-Human Influenza A(H7N2) Transmission. J Infect Dis. 2019;219:1685–1687. doi: 10.1093/infdis/jiy596 |
| 8. | Hatta M, Zhong G, Gao Y, et al. Characterization of a Feline Influenza A(H7N2) Virus. Emerg Infect Dis. 2018;24:75–86. doi: 10.3201/eid2401.171240 |
| 9. | Newman A. Hazmat Suits and 500 Shelter Cats: Rare Flu Forces New York Quarantine. The New York Times. 2017. Accessed December 01, 2017. https://www.nytimes.com/2017/01/12/nyregion/sick-cats-virus-quarantine-center-aspca.html |
| 10. | Lycett SJ, Duchatel F, Digard P. A Brief History of Bird Flu. Philos Trans R Soc Lond B Biol Sci. 2019;374:20180257. doi: 10.1098/rstb.2018.0257 |
| 11. | World Health Organization. Influenza (Avian and other zoonotic) 2018. Accessed May 29, 2023. https://www.who.int/news-room/fact-sheets/detail/influenza-(avian-and-other-zoonotic) |
| 12. | Shi J, Zeng X, Cui P, Yan C, Chen H. Alarming Situation of Emerging H5 and H7 Avian Influenza and Effective Control Strategies. Emerg Microbes Infect. n.d.;12:2155072. doi: 10.1080/22221751.2022.2155072 |
| 13. | Kawaoka Y, Webster RG. Sequence Requirements for Cleavage Activation of Influenza Virus Hemagglutinin Expressed in Mammalian Cells. Proc Natl Acad Sci. 1988;85:324–328. doi: 10.1073/pnas.85.2.324 |
| 14. | Gao R, Cao B, Hu Y, et al. Human Infection with a Novel Avian-Origin Influenza A (H7N9) Virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459 |
| 15. | Ostrowsky B, Huang A, Terry W, et al. Low Pathogenic Avian Influenza A (H7N2) Virus Infection in Immunocompromised Adult, New York, USA, 2003. Emerg Infect Dis. 2012;18:1128–1131. doi: 10.3201/eid1807.111913 |
| 16. | Terebuh P, Adija A, Edwards L, et al. Human Infection with Avian Influenza A(H7N2) Virus—Virginia, 2002. Influen Other Respir Viruses. 2018;12:529–532. doi: 10.1111/irv.12546 |
| 17. | Senne DA, Pedersen JC, Panigrahy B. Live-Bird Markets in the Northeastern United States: A Source of Avian Influenza in Commercial Poultry. In: Schrijver RS, Koch G, eds. Avian Influenza, vol. 8. Springer Netherlands; 2005:19–24. |
| 18. | Akey BL. Low-Pathogenicity H7N2 Avian Influenza Outbreak in Virginia During 2002. Avdi. 2003;47:1099–1103. doi: 10.1637/0005-2086-47.s3.1099 |
| 19. | Henzler DJ, Kradel DC, Davison S, et al. Epidemiology, Production Losses, and Control Measures Associated with an Outbreak of Avian Influenza Subtype H7N2 in Pennsylvania (1996–98). Avdi. 2003;47:1022–1036. doi: 10.1637/0005-2086-47.s3.1022 |
| 20. | Trock SC, Huntley JP. Surveillance and Control of Avian Influenza in the New York Live Bird Markets. Avdi. 2010;54:340–344. doi: 10.1637/8728-032409-ResNote.1 |
| 21. | Newbury S, Godhardt-Cooper J, Poulsen KP, Cigel F, Balanoff L, Toohey-Kurth K. Prolonged Intermittent Virus Shedding during an Outbreak of Canine Influenza A H3N2 Virus Infection in Dogs in Three Chicago Area Shelters: 16 Cases (March to May 2015). J Am Vet Med Assoc. 2016;248:1022–1026. doi: 10.2460/javma.248.9.1022 |
| 22. | Voorhees IEH, Glaser AL, Toohey-Kurth K, et al. Spread of Canine Influenza A(H3N2) Virus, United States. Emerg Infect Dis. 2017;23:1950–1957. doi: 10.3201/eid2312.170246 |
| 23. | Voorhees IEH, Dalziel BD, Glaser A, et al. Multiple Incursions and Recurrent Epidemic Fade-Out of H3N2 Canine Influenza A Virus in the United States. J Virol. 2018;92:1–15. doi: 10.1128/JVI.00323-18 |
| 24. | Jeoung H-Y, Lim S-I, Shin B-H, et al. A Novel Canine Influenza H3N2 Virus Isolated from Cats in an Animal Shelter. Vet Microbiol. 2013;165:281–286. doi: 10.1016/j.vetmic.2013.03.021 |
| 25. | De Tar L, Doyle E, O’Quin J, et al. The Guidelines for Standards of Care in Animal Shelters: Second Edition. J Shelter Med Community Anim Health. 2022;1:1–76. doi: 10.56771/ASVguidelines.2022 |
| 26. | NYC Health. Reporting Animal Diseases. 2016. Accessed Aug 2, 2023. https://web.archive.org/web/20160320154617/https://www.nyc.gov/site/doh/providers/reporting-and-services/reporting-animal-diseases.page |
| 27. | NYC Health. Reporting Animal Diseases. n.d. Accessed Jun 11, 2023. https://www.nyc.gov/site/doh/providers/reporting-and-services/reporting-animal-diseases.page |
| 28. | Mahony JB, Petrich A, Smieja M. Molecular Diagnosis of Respiratory Virus Infections. Crit Rev Clin Lab Sci. 2011;48:217–249. doi: 10.3109/10408363.2011.640976 |
| 29. | Watson CE, Bell C, Toohey-Kurth K. H3N2 Canine Influenza Virus Infection in a Dog. Vet Pathol. 2017;54:527–530. doi: 10.1177/0300985816681411 |
| 30. | Dubovi EJ. Canine Influenza. Vet Clin North Am Small Anim Pract. 2010;40:1063–1071. doi: 10.1016/j.cvsm.2010.07.005 |
| 31. | Shao W, Li X, Goraya MU, Wang S, Chen J-L. Evolution of Influenza A Virus by Mutation and Re-Assortment. Int J Mol Sci. 2017;18:1650. doi: 10.3390/ijms18081650 |
| 32. | Borland S, Gracieux P, Jones M, Mallet F, Yugueros-Marcos J. Influenza A Virus Infection in Cats and Dogs: A Literature Review in the Light of the “One Health” Concept. Front Public Health. 2020;8:83. doi: 10.3389/fpubh.2020.00083 |
| 33. | Zhao J, He W, Lu M, He H, Lai A. Emergence and Characterization of a Novel Reassortant Canine Influenza Virus Isolated from Cats. Pathogens. 2021;10:1320. doi: 10.3390/pathogens10101320 |